#
Dr. M. Baron, Statistical Machine Learning class, STAT-427/627
# BAGGING, BOOSTING, and RANDOM FORESTS
1. Bagging and Random Forests
Recall that bagging is a random forest with m = p. So, here is just
an example of a random forest.
> library(randomForest)
> rf = randomForest(mpg ~
.-name, data=Auto)†††††††††††††††††††††† # By default, m = p/3. But we can also
choose our own m.
> rf†††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††† # (m is the number of X-variables sampled
at each node)
Call:
†randomForest(formula = mpg ~ . - name, data = Auto, subset
= Z)
††††††††††††††
Type of random forest: regression
†††††††††††††††††††† Number of trees: 500
No. of variables tried at each split: 2
†††††††††
Mean of squared residuals: 8.130213
††††††††††††††††††† % Var explained: 86.53
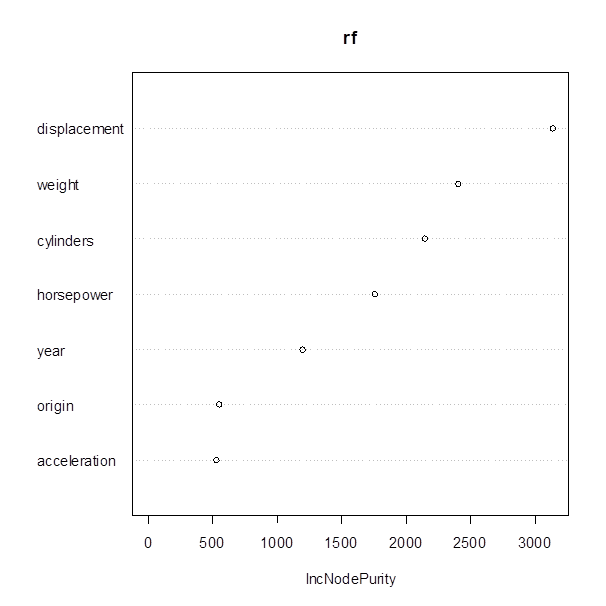
> importance(rf)††††††††††††††††††††††††††††††††††††††††††††††††††††† # Measures reduction of the nodeís impurity
(diversity), if split by the given X-variable
†††††††††††† IncNodePurity†††††††††
cylinders††††††† 2142.2611
displacement†††† 3135.1382
horsepower†††††† 1752.5234
weight†††††††††† 2399.9452
acceleration††††† 524.3148
year†††††††††††† 1196.4857
origin††††††††††† 546.0370
> varImpPlot(rf)
2. Cross-validation
> Z = sample(n,200)
> rf = randomForest(mpg
~ .-name, data=Auto, subset=Z)
> Yhat = predict(rf,
newdata=Auto[-Z,])
> mean((Yhat -
mpg[-Z])^2)
[1] 10.19022
# The mean-square error of prediction, estimated by the validation
set cross-validation, is 10.19022.
3. Searching for the optimal solution
> dim(Auto)†††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††
[1] 392††
9 ††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††
# There are 9 variables overall in the data set, minus mpg and name =
7 variables. Letís sample m = root of 7, rounded = 3.
> rf3 = randomForest(mpg
~ .-name, data=Auto, mtry=3)†††††††††††††††††† # mtry is m, the
number of X-variables available at each node
> plot(rf3)
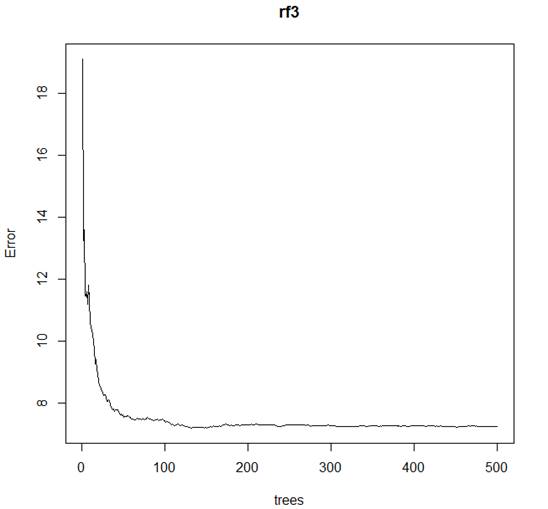
# How many trees to grow? The default is 500, but error is rather
flat after 100.
# Random forest tool has multiple output:
> names(rf3)
†[1]
"call"†††††††††††
"type"†††††††††††
"predicted"†††††† "mse"†††††††††††
†[5]
"rsq"†††††††††††† "oob.times"†††††† "importance"††††† "importanceSD"††
†[9]
"localImportance"
"proximity"†††††† "ntree"††††††††††
"mtry"††††††††††
[13] "forest"††††††††† "coefs"†††††††††† "y"†††††††††††††† "test"††††††††††
[17] "inbag"†††††††††† "terms"†††††††††
# We would like to minimize the mean squared error and to maximize R2,
the percent of total variation explained by the forest.
> which.min(rf3$mse)
[1] 147
> which.max(rf3$rsq)
[1] 147
# Alright, letís use 147 trees whose results will get averaged in
this random forest.
> rf3.147 = randomForest(mpg
~ .-name, data=Auto, mtry=3, ntree=147)
> rf3.147
Call:
†randomForest(formula = mpg ~ . - name, data = Auto, mtry = 3,††††† ntree = 147)
††††††††††††††
Type of random forest: regression
†††††††††††††††††††† Number of trees: 147
No. of variables tried at each split: 3
†††††††††
Mean of squared residuals: 7.322948
†††††† ††††††††% Var explained: 87.63
# This is an improvement in both MSE and R2, comparing
with our first random forest.
# We can optimize both m and number of trees, by cross-validation.
> Z = sample(n,n-50)†††††††††††††††††††††††††††††††††††††††††††††††††††††††††††† # Iím choosing a small test set to make the
training set close to the whole data set
> cv.err = rep(0,7)††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††† # The optimal random forest may be too
dependent on the sample size
> n.trees =
rep(0,7)
> for (m in 1:7){
+ rf.m = randomForest( mpg ~ .-name, data=Auto[Z,], mtry=m )
+ opt.trees = which.min(rf.m$mse)
+ rf.m = randomForest( mpg ~ .-name, data=Auto[Z,], mtry=m, ntree=opt.trees
)
+ Yhat = predict( rf.m, newdata=Auto[-Z,] )
+ mse = mean( (Yhat - mpg[-Z])^2 )
+ cv.err[m] = mse
+ n.trees[m] = opt.trees
+ }
> which.min(cv.err)
[1] 7
# 7? Apparently, bagging (m=p=7) was the best choice among random
forests.
> plot(cv.err);
lines(cv.err)
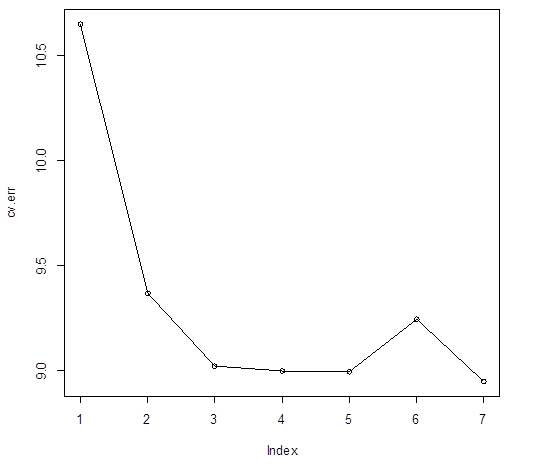
> cv.err
[1] 10.652190†
9.368370† 9.023726† 9.000002†
8.996304† 9.248892† 8.951198
> n.trees
[1] 112 494 318 208 484 319 293
# Result: here is the optimal random forest, which happened to reduce to bagging.
> rf.optimal = randomForest( mpg ~ .-name, data=Auto, mtry=7,
ntree=293 )
> rf.optimal
††††††††††††††
Type of random forest: regression
†††††††††††††††††††† Number of trees: 293
No. of variables tried at each split: 7
†††††††††
Mean of squared residuals: 7.407668
††††††††††††††††††† % Var explained: 87.81
> importance(rf.optimal)
††††††††††††
IncNodePurity
cylinders†††††††
4820.4219
displacement††††
7672.7643
horsepower††††††
2838.2213
weight††††††††††
4603.0577
acceleration†††††
647.7559
year††††††††††††
2861.6324
origin†††††††††††
133.9062